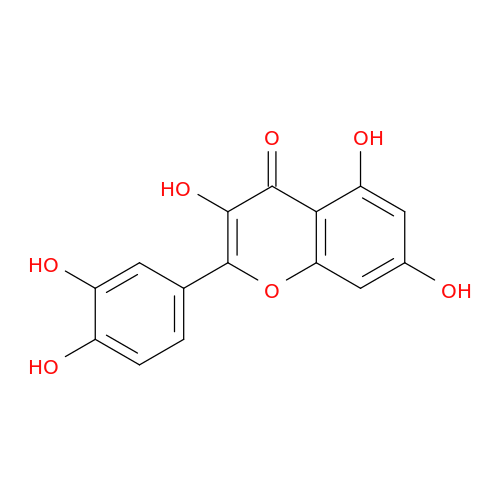
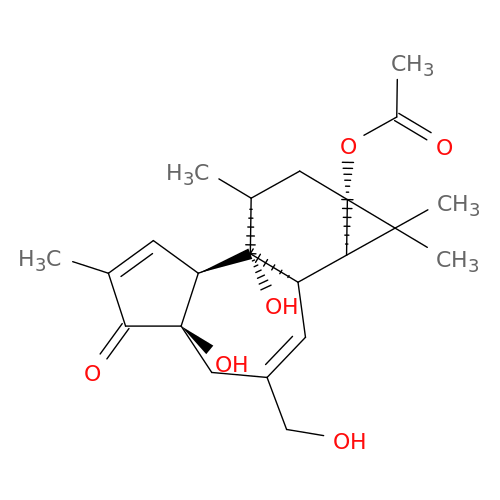
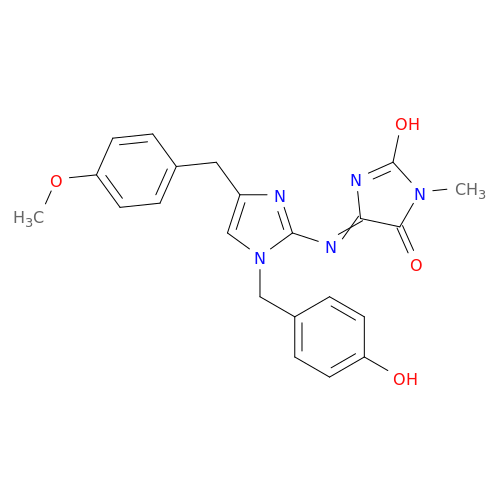
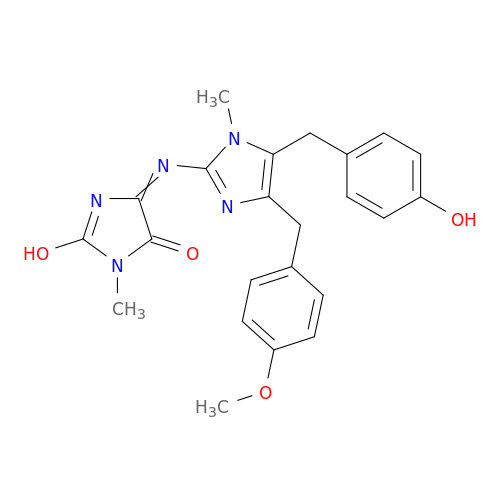
| Biological Process |
GO:0000186 |
activation of MAPKK activity |
| Biological Process |
GO:0048143 |
astrocyte activation |
| Biological Process |
GO:0098609 |
cell-cell adhesion |
| Biological Process |
GO:0030154 |
cell differentiation |
| Biological Process |
GO:0000902 |
cell morphogenesis |
| Biological Process |
GO:0007166 |
cell surface receptor signaling pathway |
| Biological Process |
GO:0071230 |
cellular response to amino acid stimulus |
| Biological Process |
GO:0071276 |
cellular response to cadmium ion |
| Biological Process |
GO:0071549 |
cellular response to dexamethasone stimulus |
| Biological Process |
GO:0035690 |
cellular response to drug |
| Biological Process |
GO:0071364 |
cellular response to epidermal growth factor stimulus |
| Biological Process |
GO:0071392 |
cellular response to estradiol stimulus |
| Biological Process |
GO:0071363 |
cellular response to growth factor stimulus |
| Biological Process |
GO:0071260 |
cellular response to mechanical stimulus |
| Biological Process |
GO:0034614 |
cellular response to reactive oxygen species |
| Biological Process |
GO:0021795 |
cerebral cortex cell migration |
| Biological Process |
GO:0007623 |
circadian rhythm |
| Biological Process |
GO:0048546 |
digestive tract morphogenesis |
| Biological Process |
GO:0016101 |
diterpenoid metabolic process |
| Biological Process |
GO:0001892 |
embryonic placenta development |
| Biological Process |
GO:0007173 |
epidermal growth factor receptor signaling pathway |
| Biological Process |
GO:0008544 |
epidermis development |
| Biological Process |
GO:0061029 |
eyelid development in camera-type eye |
| Biological Process |
GO:0001942 |
hair follicle development |
| Biological Process |
GO:0042743 |
hydrogen peroxide metabolic process |
| Biological Process |
GO:0035556 |
intracellular signal transduction |
| Biological Process |
GO:0007611 |
learning or memory |
| Biological Process |
GO:0097421 |
liver regeneration |
| Biological Process |
GO:0030324 |
lung development |
| Biological Process |
GO:0010960 |
magnesium ion homeostasis |
| Biological Process |
GO:0007494 |
midgut development |
| Biological Process |
GO:0060571 |
morphogenesis of an epithelial fold |
| Biological Process |
GO:0007275 |
multicellular organism development |
| Biological Process |
GO:0043066 |
negative regulation of apoptotic process |
| Biological Process |
GO:1905208 |
negative regulation of cardiocyte differentiation |
| Biological Process |
GO:0045930 |
negative regulation of mitotic cell cycle |
| Biological Process |
GO:0042177 |
negative regulation of protein catabolic process |
| Biological Process |
GO:0048812 |
neuron projection morphogenesis |
| Biological Process |
GO:0042698 |
ovulation cycle |
| Biological Process |
GO:0038083 |
peptidyl-tyrosine autophosphorylation |
| Biological Process |
GO:0018108 |
peptidyl-tyrosine phosphorylation |
| Biological Process |
GO:0097755 |
positive regulation of blood vessel diameter |
| Biological Process |
GO:0045780 |
positive regulation of bone resorption |
| Biological Process |
GO:0090263 |
positive regulation of canonical Wnt signaling pathway |
| Biological Process |
GO:0030307 |
positive regulation of cell growth |
| Biological Process |
GO:0030335 |
positive regulation of cell migration |
| Biological Process |
GO:0008284 |
positive regulation of cell population proliferation |
| Biological Process |
GO:0045737 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity |
| Biological Process |
GO:0045739 |
positive regulation of DNA repair |
| Biological Process |
GO:0045740 |
positive regulation of DNA replication |
| Biological Process |
GO:0050679 |
positive regulation of epithelial cell proliferation |
| Biological Process |
GO:0070374 |
positive regulation of ERK1 and ERK2 cascade |
| Biological Process |
GO:0048146 |
positive regulation of fibroblast proliferation |
| Biological Process |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle |
| Biological Process |
GO:0050729 |
positive regulation of inflammatory response |
| Biological Process |
GO:0033674 |
positive regulation of kinase activity |
| Biological Process |
GO:0043406 |
positive regulation of MAP kinase activity |
| Biological Process |
GO:1901224 |
positive regulation of NIK/NF-kappaB signaling |
| Biological Process |
GO:0010750 |
positive regulation of nitric oxide mediated signal transduction |
| Biological Process |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation |
| Biological Process |
GO:0042327 |
positive regulation of phosphorylation |
| Biological Process |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA |
| Biological Process |
GO:1902722 |
positive regulation of prolactin secretion |
| Biological Process |
GO:0051897 |
positive regulation of protein kinase B signaling |
| Biological Process |
GO:1900020 |
positive regulation of protein kinase C activity |
| Biological Process |
GO:1902966 |
positive regulation of protein localization to early endosome |
| Biological Process |
GO:1903078 |
positive regulation of protein localization to plasma membrane |
| Biological Process |
GO:0001934 |
positive regulation of protein phosphorylation |
| Biological Process |
GO:0048661 |
positive regulation of smooth muscle cell proliferation |
| Biological Process |
GO:0032930 |
positive regulation of superoxide anion generation |
| Biological Process |
GO:0051968 |
positive regulation of synaptic transmission, glutamatergic |
| Biological Process |
GO:0045893 |
positive regulation of transcription, DNA-templated |
| Biological Process |
GO:0045944 |
positive regulation of transcription by RNA polymerase II |
| Biological Process |
GO:0045907 |
positive regulation of vasoconstriction |
| Biological Process |
GO:0046777 |
protein autophosphorylation |
| Biological Process |
GO:0042127 |
regulation of cell population proliferation |
| Biological Process |
GO:0070372 |
regulation of ERK1 and ERK2 cascade |
| Biological Process |
GO:0046328 |
regulation of JNK cascade |
| Biological Process |
GO:0050999 |
regulation of nitric-oxide synthase activity |
| Biological Process |
GO:0050730 |
regulation of peptidyl-tyrosine phosphorylation |
| Biological Process |
GO:0014066 |
regulation of phosphatidylinositol 3-kinase signaling |
| Biological Process |
GO:0051592 |
response to calcium ion |
| Biological Process |
GO:0033590 |
response to cobalamin |
| Biological Process |
GO:0033594 |
response to hydroxyisoflavone |
| Biological Process |
GO:0006970 |
response to osmotic stress |
| Biological Process |
GO:0070141 |
response to UV-A |
| Biological Process |
GO:0007435 |
salivary gland morphogenesis |
| Biological Process |
GO:0007165 |
signal transduction |
| Biological Process |
GO:0043586 |
tongue development |
| Biological Process |
GO:0006412 |
translation |
| Biological Process |
GO:0007169 |
transmembrane receptor protein tyrosine kinase signaling pathway |
| Biological Process |
GO:0042060 |
wound healing |
| molecular function |
GO:0051015 |
actin filament binding |
| molecular function |
GO:0005524 |
ATP binding |
| molecular function |
GO:0005516 |
calmodulin binding |
| molecular function |
GO:0003682 |
chromatin binding |
| molecular function |
GO:0019899 |
enzyme binding |
| molecular function |
GO:0005006 |
epidermal growth factor-activated receptor activity |
| molecular function |
GO:0048408 |
epidermal growth factor binding |
| molecular function |
GO:0042802 |
identical protein binding |
| molecular function |
GO:0005178 |
integrin binding |
| molecular function |
GO:0016301 |
kinase activity |
| molecular function |
GO:0030235 |
nitric-oxide synthase regulator activity |
| molecular function |
GO:0044877 |
protein-containing complex binding |
| molecular function |
GO:0019901 |
protein kinase binding |
| molecular function |
GO:0019903 |
protein phosphatase binding |
| molecular function |
GO:0004713 |
protein tyrosine kinase activity |
| molecular function |
GO:0005102 |
signaling receptor binding |
| molecular function |
GO:0004714 |
transmembrane receptor protein tyrosine kinase activity |
| molecular function |
GO:0004888 |
transmembrane signaling receptor activity |
| molecular function |
GO:0031625 |
ubiquitin protein ligase binding |
| cellular component |
GO:0016324 |
apical plasma membrane |
| cellular component |
GO:0009925 |
basal plasma membrane |
| cellular component |
GO:0016323 |
basolateral plasma membrane |
| cellular component |
GO:0030054 |
cell junction |
| cellular component |
GO:0009986 |
cell surface |
| cellular component |
GO:0005737 |
cytoplasm |
| cellular component |
GO:0031901 |
early endosome membrane |
| cellular component |
GO:0030139 |
endocytic vesicle |
| cellular component |
GO:0005789 |
endoplasmic reticulum membrane |
| cellular component |
GO:0005768 |
endosome |
| cellular component |
GO:0010008 |
endosome membrane |
| cellular component |
GO:0000139 |
Golgi membrane |
| cellular component |
GO:0005887 |
integral component of plasma membrane |
| cellular component |
GO:0016020 |
membrane |
| cellular component |
GO:0045121 |
membrane raft |
| cellular component |
GO:0097489 |
multivesicular body, internal vesicle lumen |
| cellular component |
GO:0031965 |
nuclear membrane |
| cellular component |
GO:0005634 |
nucleus |
| cellular component |
GO:0048471 |
perinuclear region of cytoplasm |
| cellular component |
GO:0005886 |
plasma membrane |
| cellular component |
GO:0032991 |
protein-containing complex |
| cellular component |
GO:0043235 |
receptor complex |
| cellular component |
GO:0045202 |
synapse |