Showing entry for Anthranilic acid
| General Compound Information | |
|---|---|
| BXGC Id | BXGC0000539 |
| Compound Name | Anthranilic acid |
| Structure | 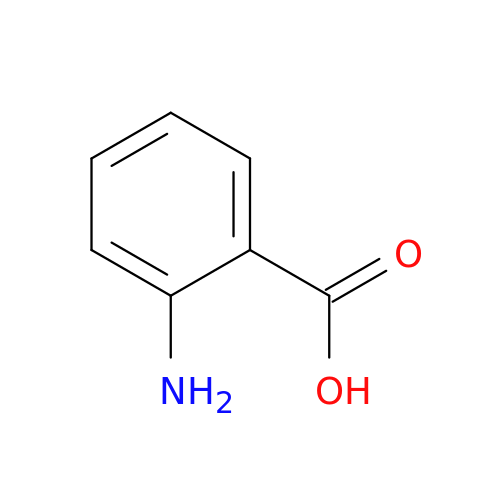 |
| Formula | C7H7NO2 |
| InchiKey | RWZYAGGXGHYGMB-UHFFFAOYSA-N |
| SMILES | NC1=CC=CC=C1C(O)=O |
| Inchi | InChI=1S/C7H7NO2/c8-6-4-2-1-3-5(6)7(9)10/h1-4H,8H2,(H,9,10) |
| IUPAC | 2-aminobenzoic acid |
| Molecular Weight | 137.14 |
| Pubchem Id | 227 |
| Chembl Id | CHEMBL14173 |
| Targets of Information Source | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank | DB04166 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB | BE2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Binding DB | 50376751 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CHEMBL | CHEMBL14173 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Foods containing the compound | |||||||||||||||||||||||||||||||||||||
| Foods |
|
||||||||||||||||||||||||||||||||||||
| The compound-realted diseases | ||||||
| Diseases |
|
|||||