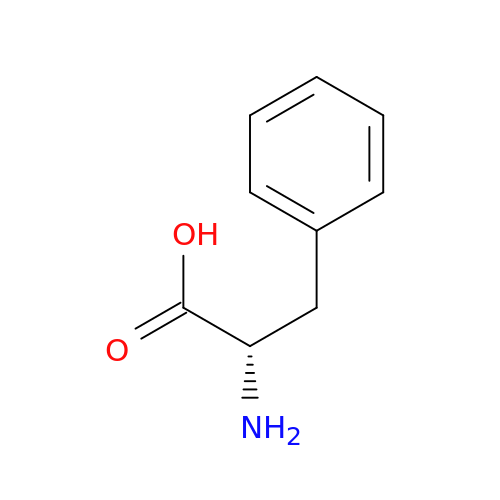
| P0C061 |
BXGT007382 |
Gramicidin S synthase 1 |
reviewed |
grsA |
- |
- |
- |
- |
- |
| P27001 |
BXGT009149 |
Phenylalanine--tRNA ligase alpha subunit |
reviewed |
pheS |
- |
- |
- |
- |
- |
| Q59771 |
BXGT015265 |
Phenylalanine dehydrogenase |
reviewed |
pdh |
- |
- |
- |
- |
- |
| P80561 |
BXGT011917 |
Aminopeptidase S |
reviewed |
SGR_5809 |
- |
12700000 |
- |
- |
- |
| Q9QWW1 |
BXGT021381 |
Homer protein homolog 2 |
reviewed |
Homer2 |
- |
- |
- |
- |
- |
| P70552 |
BXGT011674 |
GTP cyclohydrolase 1 feedback regulatory protein |
reviewed |
Gchfr |
- |
- |
- |
- |
- |
| P0AB91 |
BXGT007223 |
Phospho-2-dehydro-3-deoxyheptonate aldolase, Phe-sensitive |
reviewed |
aroG |
- |
- |
- |
- |
- |
| P32449 |
BXGT009639 |
Phospho-2-dehydro-3-deoxyheptonate aldolase, tyrosine-inhibited |
reviewed |
ARO4 |
- |
- |
- |
- |
- |
| Q6STF1 |
BXGT016527 |
L-amino-acid oxidase |
reviewed |
|
- |
- |
- |
- |
- |
| P08254 |
BXGT006758 |
Stromelysin-1 |
reviewed |
MMP3 |
Enzyme |
- |
- |
- |
- |
| P04816 |
BXGT006268 |
Leucine-specific-binding protein |
reviewed |
livK |
- |
- |
- |
180 |
- |
| Q16773 |
BXGT013667 |
Kynurenine--oxoglutarate transaminase 1 |
reviewed |
KYAT1 |
Enzyme |
- |
- |
- |
- |
| P96084 |
BXGT012167 |
Proline iminopeptidase |
reviewed |
pip |
- |
- |
- |
- |
- |
| P05187 |
BXGT006339 |
Alkaline phosphatase, placental type |
reviewed |
ALPP |
Enzyme |
- |
- |
- |
- |
| P00772 |
BXGT005648 |
Chymotrypsin-like elastase family member 1 |
reviewed |
CELA1 |
- |
- |
- |
- |
- |
| P01180 |
BXGT005741 |
Vasopressin-neurophysin 2-copeptin |
reviewed |
AVP |
- |
- |
- |
- |
- |
| Q9LCC8 |
BXGT021067 |
D-Amino acid amidase |
unreviewed |
daaA |
- |
- |
- |
- |
- |
| P00918 |
BXGT005690 |
Carbonic anhydrase 2 |
reviewed |
CA2 |
- |
- |
- |
- |
- |
| P01175 |
BXGT005740 |
Oxytocin-neurophysin 1 |
reviewed |
OXT |
- |
- |
- |
- |
- |
| Q9YBQ2 |
BXGT022365 |
Acylamino-acid-releasing enzyme |
reviewed |
APE_1547.1 |
- |
- |
- |
- |
- |
| P81382 |
BXGT011946 |
L-amino-acid oxidase |
reviewed |
|
- |
- |
- |
- |
- |
| Q8VPD4 |
BXGT018940 |
L-amino acid oxidase |
reviewed |
|
- |
- |
- |
- |
- |
| Q8KBW6 |
BXGT018410 |
Prephenate dehydratase |
unreviewed |
pheA |
- |
- |
- |
- |
- |
| P96896 |
BXGT012192 |
Leucine-responsive regulatory protein |
unreviewed |
lrp |
- |
- |
- |
- |
- |
| O53512 |
BXGT004702 |
Phospho-2-dehydro-3-deoxyheptonate aldolase AroG |
reviewed |
aroG |
- |
- |
- |
21000 |
- |
| P0A8P1 |
BXGT007151 |
Leucyl/phenylalanyl-tRNA--protein transferase |
reviewed |
aat |
- |
- |
- |
- |
- |
| Q5W9R9 |
BXGT015899 |
Phenylalanine 2-monooxygenase precursor |
reviewed |
|
- |
- |
- |
- |
- |
| P04825 |
BXGT006271 |
Aminopeptidase N |
reviewed |
pepN |
- |
- |
- |
- |
- |
| Q9S7N2 |
BXGT021570 |
L-tryptophan--pyruvate aminotransferase 1 |
reviewed |
TAA1 |
- |
- |
- |
- |
- |
| Q9WXM1 |
BXGT021955 |
L-Ala-D/L-Glu epimerase |
reviewed |
TM_0006 |
- |
- |
- |
- |
- |
| Q16651 |
BXGT013649 |
Prostasin |
reviewed |
PRSS8 |
Enzyme |
- |
- |
- |
- |
| O53512 |
BXGT004702 |
Phospho-2-dehydro-3-deoxyheptonate aldolase AroG |
reviewed |
aroG |
- |
- |
- |
- |
- |
| Q9Y285 |
BXGT022224 |
Phenylalanine--tRNA ligase alpha subunit |
reviewed |
FARSA |
- |
- |
- |
- |
- |
| Q9KT04 |
BXGT020994 |
Uncharacterized protein |
unreviewed |
VC_1101 |
- |
- |
- |
- |
- |
| A1R118 |
BXGT001027 |
Prephenate dehydratase |
unreviewed |
pheA |
- |
- |
- |
- |
- |
| P08312 |
BXGT006763 |
Phenylalanine--tRNA ligase alpha subunit |
reviewed |
pheS |
- |
- |
- |
- |
- |
| P30967 |
BXGT009487 |
Phenylalanine-4-hydroxylase |
reviewed |
phhA |
- |
- |
- |
- |
- |
| Q9X0L9 |
BXGT022068 |
Branched chain amino acid ABC transporter, periplasmic amino acid-binding protein |
unreviewed |
TM_1135 |
- |
- |
- |
- |
- |
| Q84FL5 |
BXGT017553 |
AdmH |
unreviewed |
|
- |
- |
- |
- |
- |
| Q5M4H8 |
BXGT015569 |
Di-or tripeptide:H+ symporter |
unreviewed |
dtpT |
- |
- |
- |
- |
- |
| P14618 |
BXGT008027 |
Pyruvate kinase PKM |
reviewed |
PKM |
Kinase |
- |
- |
- |
- |
| E9P5T5 |
BXGT003186 |
Glutamate receptor 1 |
unreviewed |
|
- |
- |
- |
- |
- |
| P0A988 |
BXGT007175 |
Beta sliding clamp |
reviewed |
dnaN |
- |
- |
- |
- |
- |
| P42738 |
BXGT010336 |
Chorismate mutase 1, chloroplastic |
reviewed |
CM1 |
- |
- |
- |
- |
- |
| A7Y7W1 |
BXGT001643 |
Periplasmic dipeptide-binding protein |
unreviewed |
dppA |
- |
- |
- |
- |
- |
| P9WML5 |
BXGT012392 |
Putative phenylalanine aminotransferase |
reviewed |
pat |
- |
- |
- |
- |
- |
| Q9K169 |
BXGT020867 |
Phospho-2-dehydro-3-deoxyheptonate aldolase |
unreviewed |
aroG |
- |
- |
- |
- |
- |
| P03418 |
BXGT006066 |
Nucleoprotein |
reviewed |
N |
- |
- |
- |
- |
- |
| P00439 |
BXGT005539 |
Phenylalanine-4-hydroxylase |
reviewed |
PAH |
- |
- |
- |
- |
- |
| O95363 |
BXGT005375 |
Phenylalanine--tRNA ligase, mitochondrial |
reviewed |
FARS2 |
Enzyme |
- |
- |
- |
- |
| Q4QDU3 |
BXGT014873 |
UTP--glucose-1-phosphate uridylyltransferase |
unreviewed |
UGP |
- |
- |
- |
- |
- |
| P14779 |
BXGT008052 |
Bifunctional cytochrome P450/NADPH--P450 reductase |
reviewed |
cyp102A1 |
- |
- |
- |
- |
- |
| P25084 |
BXGT009013 |
Transcriptional activator protein LasR |
reviewed |
lasR |
- |
- |
- |
- |
- |
| Q8RY79 |
BXGT018751 |
Tyrosine decarboxylase 1 |
reviewed |
ELI5 |
- |
- |
- |
- |
- |
| P40969 |
BXGT010213 |
Centromere DNA-binding protein complex CBF3 subunit B |
reviewed |
CEP3 |
- |
- |
- |
- |
- |
| C4YJ02 |
BXGT024201 |
Aromatic amino acid aminotransferase I |
unreviewed |
CAWG_03814 |
- |
- |
- |
- |
- |
| A0A1D8PMC5 |
BXGT024202 |
Aromatic-amino-acid:2-oxoglutarate transaminase |
unreviewed |
ARO9 |
- |
- |
- |
- |
- |
| F8W4B7 |
BXGT003330 |
Histone deacetylase 6 |
unreviewed |
hdac6 |
- |
- |
- |
- |
- |
| Q2YPM5 |
BXGT025823 |
Prephenate dehydratase |
unreviewed |
pheA |
- |
- |
- |
- |
- |
| Q0P864 |
BXGT013112 |
Putative methyl-accepting chemotaxis signal transduction protein |
unreviewed |
Cj1564 |
- |
- |
- |
- |
- |
| O67854 |
BXGT005012 |
Na(+):neurotransmitter symporter (Snf family) |
unreviewed |
snf |
- |
- |
- |
- |
- |
| Q16769 |
BXGT013665 |
Glutaminyl-peptide cyclotransferase |
reviewed |
QPCT |
Enzyme |
- |
- |
- |
- |
| A0A4Y7X244 |
BXGT026014 |
Sodium-dependent transporter |
unreviewed |
E2L07_01100 |
- |
- |
- |
- |
- |
| A0A0J9X1X3 |
BXGT026057 |
L-lysine oxidase |
unreviewed |
|
- |
- |
- |
- |
- |