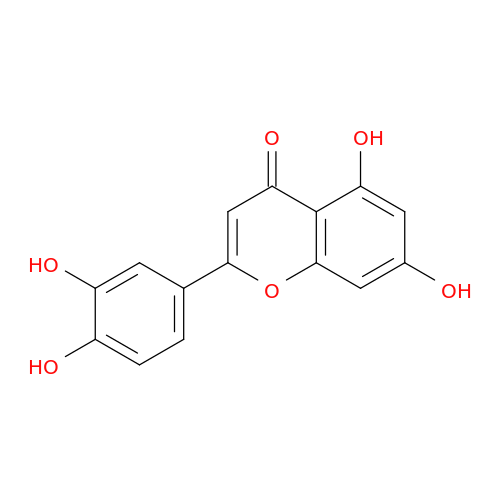
| P11309 |
BXGT007685 |
Serine/threonine-protein kinase pim-1 |
reviewed |
Kinase |
PIM1 |
Activity |
'=' |
0 |
% |
| P16116 |
BXGT008197 |
Aldo-keto reductase family 1 member B1 |
reviewed |
- |
AKR1B1 |
IC50 |
'=' |
9570 |
nM |
| B4URF0 |
BXGT002146 |
Neuraminidase |
unreviewed |
- |
NA |
IC50 |
'=' |
33700 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Km |
'=' |
2400 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Activity |
'=' |
71 |
nM/s |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Activity |
'=' |
110 |
nM/s |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Ki |
'=' |
1900 |
nM |
| P08183 |
BXGT006741 |
ATP-dependent translocase ABCB1 |
reviewed |
Transporter |
ABCB1 |
IC50 |
'=' |
19600 |
nM |
| Q14191 |
BXGT013452 |
Werner syndrome ATP-dependent helicase |
reviewed |
Enzyme |
WRN |
Potency |
- |
3040 |
nM |
| B2RXH2 |
BXGT002006 |
Lysine-specific demethylase 4E |
reviewed |
- |
KDM4E |
Potency |
'=' |
8912 |
nM |
| P10481 |
BXGT007573 |
Sialidase |
reviewed |
- |
nanH |
IC50 |
'=' |
4300 |
nM |
| Q9UBT6 |
BXGT021664 |
DNA polymerase kappa |
reviewed |
- |
POLK |
Potency |
- |
6309 |
nM |
| P16050 |
BXGT008184 |
Arachidonate 15-lipoxygenase |
reviewed |
Enzyme |
ALOX15 |
Potency |
'=' |
2511 |
nM |
| P04839 |
BXGT006273 |
Cytochrome b-245 heavy chain |
reviewed |
Ion channel |
CYBB |
Inhibition |
- |
- |
- |
| B5U2Z5 |
BXGT002179 |
Phenol oxidase |
unreviewed |
- |
|
Inhibition |
- |
- |
- |
| O42713 |
BXGT004563 |
Polyphenol oxidase 2 |
reviewed |
- |
PPO2 |
Inhibition |
'=' |
11 |
% |
| P08170 |
BXGT006736 |
Seed linoleate 13S-lipoxygenase-1 |
reviewed |
- |
LOX1.1 |
Vmax |
'=' |
0 |
uM |
| P11388 |
BXGT007697 |
DNA topoisomerase 2-alpha |
reviewed |
- |
TOP2A |
Fluorescence intensity |
'=' |
7 |
- |
| P07265 |
BXGT006583 |
Alpha-glucosidase MAL62 |
reviewed |
- |
MAL62 |
logIC50 |
'=' |
1 |
- |
| Q16678 |
BXGT013656 |
Cytochrome P450 1B1 |
reviewed |
Enzyme |
CYP1B1 |
IC50 |
'=' |
360 |
nM |
| P51450 |
BXGT010847 |
Nuclear receptor ROR-gamma |
reviewed |
- |
Rorc |
Potency |
'=' |
14125 |
nM |
| P16050 |
BXGT008184 |
Arachidonate 15-lipoxygenase |
reviewed |
Enzyme |
ALOX15 |
Potency |
'=' |
1584 |
nM |
| P04637 |
BXGT006243 |
Cellular tumor antigen p53 |
reviewed |
Transcription factor |
TP53 |
Potency |
'=' |
5011 |
nM |
| Q9WUL0 |
BXGT021941 |
DNA topoisomerase 1 |
reviewed |
- |
Top1 |
IC50 |
'=' |
5000 |
nM |
| B4URF0 |
BXGT002146 |
Neuraminidase |
unreviewed |
- |
NA |
IC50 |
'=' |
32600 |
nM |
| O95149 |
BXGT005350 |
Snurportin-1 |
reviewed |
- |
SNUPN |
Potency |
- |
44668 |
nM |
| Q14974 |
BXGT013516 |
Importin subunit beta-1 |
reviewed |
Transporter |
KPNB1 |
Potency |
- |
44668 |
nM |
| Q9UNQ0 |
BXGT021792 |
ATP-binding cassette sub-family G member 2 |
reviewed |
Transporter |
ABCG2 |
Activity |
- |
- |
- |
| P27169 |
BXGT009161 |
Serum paraoxonase/arylesterase 1 |
reviewed |
- |
PON1 |
Ka |
'=' |
1 |
10'5/M |
| P47989 |
BXGT010602 |
Xanthine dehydrogenase/oxidase |
reviewed |
Enzyme |
XDH |
Ki |
'=' |
2900 |
nM |
| P10828 |
BXGT007624 |
Thyroid hormone receptor beta |
reviewed |
Nuclear receptor |
THRB |
Potency |
'=' |
44668 |
nM |
| P03468 |
BXGT006079 |
Neuraminidase |
reviewed |
- |
NA |
IC50 |
'=' |
9 |
ug.mL-1 |
| Q03164 |
BXGT012700 |
Histone-lysine N-methyltransferase 2A |
reviewed |
- |
KMT2A |
Potency |
'=' |
31622 |
nM |
| O00255 |
BXGT003903 |
Menin |
reviewed |
- |
MEN1 |
Potency |
'=' |
31622 |
nM |
| P06276 |
BXGT006430 |
Cholinesterase |
reviewed |
- |
BCHE |
Ki |
'=' |
166100 |
nM |
| P49327 |
BXGT010694 |
Fatty acid synthase |
reviewed |
Enzyme |
FASN |
IC50 |
'=' |
62500 |
nM |
| P54132 |
BXGT011032 |
Bloom syndrome protein |
reviewed |
- |
BLM |
Potency |
'=' |
35481 |
nM |
| P00747 |
BXGT005639 |
Plasminogen |
reviewed |
Enzyme |
PLG |
Inhibition |
'=' |
28 |
% |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Kcat/Km |
'=' |
0 |
/uM/s |
| P02545 |
BXGT005889 |
Prelamin-A/C |
reviewed |
- |
LMNA |
Potency |
'=' |
3162 |
nM |
| P00915 |
BXGT005689 |
Carbonic anhydrase 1 |
reviewed |
- |
CA1 |
Ki |
'>' |
10000 |
nM |
| P18054 |
BXGT008370 |
Arachidonate 12-lipoxygenase, 12S-type |
reviewed |
Enzyme |
ALOX12 |
Potency |
'=' |
15848 |
nM |
| P15428 |
BXGT008121 |
15-hydroxyprostaglandin dehydrogenase [NAD(+)] |
reviewed |
- |
HPGD |
Potency |
'=' |
11220 |
nM |
| P08684 |
BXGT006812 |
Cytochrome P450 3A4 |
reviewed |
Enzyme |
CYP3A4 |
AC50 |
'=' |
5011 |
nM |
| Q9NUW8 |
BXGT021219 |
Tyrosyl-DNA phosphodiesterase 1 |
reviewed |
Enzyme |
TDP1 |
Potency |
- |
29092 |
nM |
| P04637 |
BXGT006243 |
Cellular tumor antigen p53 |
reviewed |
Transcription factor |
TP53 |
Potency |
- |
28183 |
nM |
| P16473 |
BXGT008234 |
Thyrotropin receptor |
reviewed |
G-protein coupled receptor |
TSHR |
Potency |
'=' |
39810 |
nM |
| Q03164 |
BXGT012700 |
Histone-lysine N-methyltransferase 2A |
reviewed |
- |
KMT2A |
Potency |
'=' |
14125 |
nM |
| O00255 |
BXGT003903 |
Menin |
reviewed |
- |
MEN1 |
Potency |
'=' |
14125 |
nM |
| Q16678 |
BXGT013656 |
Cytochrome P450 1B1 |
reviewed |
Enzyme |
CYP1B1 |
IC50 |
'=' |
79 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Kcat |
'=' |
3 |
/s |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Kcat/Km |
'=' |
0 |
/uM/s |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Activity |
'=' |
100 |
nM/s |
| P10636 |
BXGT007596 |
Microtubule-associated protein tau |
reviewed |
- |
MAPT |
Potency |
'=' |
12589 |
nM |
| Q9NPH5 |
BXGT021159 |
NADPH oxidase 4 |
reviewed |
Enzyme |
NOX4 |
Inhibition |
'=' |
106 |
% |
| P05177 |
BXGT006336 |
Cytochrome P450 1A2 |
reviewed |
Enzyme |
CYP1A2 |
IC50 |
'=' |
3370 |
nM |
| B4URF0 |
BXGT002146 |
Neuraminidase |
unreviewed |
- |
NA |
IC50 |
'=' |
53300 |
nM |
| P12822 |
BXGT007850 |
Angiotensin-converting enzyme |
reviewed |
- |
ACE |
IC50 |
'=' |
290000 |
nM |
| P04745 |
BXGT006255 |
Alpha-amylase 1 |
reviewed |
- |
AMY1A |
IC50 |
'=' |
18400 |
nM |
| P28907 |
BXGT009301 |
ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 |
reviewed |
Enzyme |
CD38 |
IC50 |
'=' |
8200 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Km |
'=' |
53000 |
nM |
| P24941 |
BXGT009002 |
Cyclin-dependent kinase 2 |
reviewed |
Kinase |
CDK2 |
IC50 |
'=' |
20000 |
nM |
| P47989 |
BXGT010602 |
Xanthine dehydrogenase/oxidase |
reviewed |
Enzyme |
XDH |
IC50 |
'=' |
11540 |
nM |
| P15917 |
BXGT008169 |
Lethal factor |
reviewed |
- |
lef |
Potency |
'=' |
1000 |
nM |
| P54132 |
BXGT011032 |
Bloom syndrome protein |
reviewed |
- |
BLM |
Potency |
'=' |
15848 |
nM |
| P14780 |
BXGT008053 |
Matrix metalloproteinase-9 |
reviewed |
Enzyme |
MMP9 |
Inhibition |
'>' |
30 |
% |
| P28839 |
BXGT009296 |
Cytosol aminopeptidase |
reviewed |
- |
LAP3 |
IC50 |
'=' |
49600 |
nM |
| Q9HC97 |
BXGT020527 |
G-protein coupled receptor 35 |
reviewed |
G-protein coupled receptor |
GPR35 |
IC50 |
'=' |
18600 |
nM |
| Q04760 |
BXGT012782 |
Lactoylglutathione lyase |
reviewed |
- |
GLO1 |
IC50 |
'=' |
7700 |
nM |
| O42713 |
BXGT004563 |
Polyphenol oxidase 2 |
reviewed |
- |
PPO2 |
Inhibition |
'=' |
17 |
% |
| P27169 |
BXGT009161 |
Serum paraoxonase/arylesterase 1 |
reviewed |
- |
PON1 |
Kq |
'=' |
1 |
10'12/M/s |
| P11388 |
BXGT007697 |
DNA topoisomerase 2-alpha |
reviewed |
- |
TOP2A |
Fluorescence intensity |
'=' |
133 |
% |
| P08684 |
BXGT006812 |
Cytochrome P450 3A4 |
reviewed |
Enzyme |
CYP3A4 |
Potency |
'=' |
5011 |
nM |
| Q9UBT6 |
BXGT021664 |
DNA polymerase kappa |
reviewed |
- |
POLK |
Potency |
- |
891 |
nM |
| P00352 |
BXGT005510 |
Retinal dehydrogenase 1 |
reviewed |
Enzyme |
ALDH1A1 |
Potency |
'=' |
19952 |
nM |
| P54132 |
BXGT011032 |
Bloom syndrome protein |
reviewed |
- |
BLM |
Potency |
'=' |
19952 |
nM |
| P46063 |
BXGT010501 |
ATP-dependent DNA helicase Q1 |
reviewed |
Enzyme |
RECQL |
Potency |
'=' |
39810 |
nM |
| P08659 |
BXGT006805 |
Luciferin 4-monooxygenase |
reviewed |
- |
|
Potency |
- |
23934 |
nM |
| O42713 |
BXGT004563 |
Polyphenol oxidase 2 |
reviewed |
- |
PPO2 |
Inhibition |
'=' |
9 |
% |
| Q9XUB2 |
BXGT022194 |
Zinc finger protein mex-5 |
reviewed |
- |
mex-5 |
EC50 |
'=' |
31000 |
nM |
| Q96KQ7 |
BXGT019687 |
Histone-lysine N-methyltransferase EHMT2 |
reviewed |
Epigenetic regulator |
EHMT2 |
Potency |
- |
10000 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Kcat |
'=' |
6 |
/s |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Km |
'=' |
31000 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Km |
'=' |
43000 |
nM |
| P08684 |
BXGT006812 |
Cytochrome P450 3A4 |
reviewed |
Enzyme |
CYP3A4 |
Potency |
'=' |
15848 |
nM |
| P11387 |
BXGT007696 |
DNA topoisomerase 1 |
reviewed |
Enzyme |
TOP1 |
Fluorescence intensity |
'=' |
0 |
- |
| P11387 |
BXGT007696 |
DNA topoisomerase 1 |
reviewed |
Enzyme |
TOP1 |
Fluorescence intensity |
'=' |
0 |
- |
| Q965D7 |
BXGT019602 |
Beta-hydroxyacyl-ACP dehydratase |
unreviewed |
- |
fabZ |
Ki |
'=' |
11500 |
nM |
| Q965D5 |
BXGT019600 |
Enoyl-acyl-carrier protein reductase |
unreviewed |
- |
fabI |
Ki |
'=' |
2100 |
nM |
| Q965D6 |
BXGT019601 |
3-oxoacyl-acyl-carrier protein reductase |
unreviewed |
- |
fabG |
IC50 |
'=' |
4000 |
nM |
| B2RXH2 |
BXGT002006 |
Lysine-specific demethylase 4E |
reviewed |
- |
KDM4E |
Potency |
'=' |
12589 |
nM |
| P21397 |
BXGT008644 |
Amine oxidase [flavin-containing] A |
reviewed |
- |
MAOA |
IC50 |
'=' |
4900 |
nM |
| Q9Y253 |
BXGT022216 |
DNA polymerase eta |
reviewed |
- |
POLH |
Potency |
- |
3162 |
nM |
| O60218 |
BXGT004858 |
Aldo-keto reductase family 1 member B10 |
reviewed |
Enzyme |
AKR1B10 |
Inhibition |
'=' |
33 |
% |
| P08170 |
BXGT006736 |
Seed linoleate 13S-lipoxygenase-1 |
reviewed |
- |
LOX1.1 |
IC50 |
'=' |
3200 |
nM |
| P51450 |
BXGT010847 |
Nuclear receptor ROR-gamma |
reviewed |
- |
Rorc |
Potency |
'=' |
22387 |
nM |
| O75874 |
BXGT005157 |
Isocitrate dehydrogenase [NADP] cytoplasmic |
reviewed |
- |
IDH1 |
Activity |
'=' |
93 |
% |
| P15917 |
BXGT008169 |
Lethal factor |
reviewed |
- |
lef |
Potency |
'=' |
25118 |
nM |
| Q8IEK1 |
BXGT018263 |
M1-family alanyl aminopeptidase |
unreviewed |
- |
PF3D7_1311800 |
IC50 |
'=' |
22860 |
nM |
| P25779 |
BXGT009067 |
Cruzipain |
reviewed |
- |
|
IC50 |
'=' |
27300 |
nM |
| P08170 |
BXGT006736 |
Seed linoleate 13S-lipoxygenase-1 |
reviewed |
- |
LOX1.1 |
Activity |
- |
- |
- |
| P08170 |
BXGT006736 |
Seed linoleate 13S-lipoxygenase-1 |
reviewed |
- |
LOX1.1 |
Km |
'=' |
189600 |
nM |
| P11309 |
BXGT007685 |
Serine/threonine-protein kinase pim-1 |
reviewed |
Kinase |
PIM1 |
#NAME? |
'=' |
0 |
uM |
| P25099 |
BXGT009016 |
Adenosine receptor A1 |
reviewed |
- |
Adora1 |
Ki |
'=' |
1660 |
nM |
| P25099 |
BXGT009016 |
Adenosine receptor A1 |
reviewed |
- |
Adora1 |
Ki |
'=' |
1550 |
nM |
| P04062 |
BXGT006140 |
Lysosomal acid glucosylceramidase |
reviewed |
- |
GBA |
Potency |
- |
28183 |
nM |
| P46063 |
BXGT010501 |
ATP-dependent DNA helicase Q1 |
reviewed |
Enzyme |
RECQL |
Potency |
'=' |
12589 |
nM |
| O95067 |
BXGT005347 |
G2/mitotic-specific cyclin-B2 |
reviewed |
Enzyme modulator |
CCNB2 |
IC50 |
'=' |
6200 |
nM |
| Q8WWL7 |
BXGT019000 |
G2/mitotic-specific cyclin-B3 |
reviewed |
Enzyme modulator |
CCNB3 |
IC50 |
'=' |
6200 |
nM |
| P14635 |
BXGT008031 |
G2/mitotic-specific cyclin-B1 |
reviewed |
Enzyme modulator |
CCNB1 |
IC50 |
'=' |
6200 |
nM |
| P06493 |
BXGT006445 |
Cyclin-dependent kinase 1 |
reviewed |
Kinase |
CDK1 |
IC50 |
'=' |
6200 |
nM |
| P35354 |
BXGT009816 |
Prostaglandin G/H synthase 2 |
reviewed |
Enzyme |
PTGS2 |
Inhibition |
'=' |
80 |
% |
| Q965D6 |
BXGT019601 |
3-oxoacyl-acyl-carrier protein reductase |
unreviewed |
- |
fabG |
Ki |
'=' |
800 |
nM |
| Q9NPH5 |
BXGT021159 |
NADPH oxidase 4 |
reviewed |
Enzyme |
NOX4 |
Inhibition |
'=' |
110 |
% |
| O75164 |
BXGT005104 |
Lysine-specific demethylase 4A |
reviewed |
- |
KDM4A |
Potency |
- |
44668 |
nM |
| B4URF0 |
BXGT002146 |
Neuraminidase |
unreviewed |
- |
NA |
Ki |
'=' |
19600 |
nM |
| P07943 |
BXGT006700 |
Aldo-keto reductase family 1 member B1 |
reviewed |
- |
Akr1b1 |
IC50 |
'=' |
1540 |
nM |
| P10636 |
BXGT007596 |
Microtubule-associated protein tau |
reviewed |
- |
MAPT |
Potency |
'=' |
28183 |
nM |
| P16116 |
BXGT008197 |
Aldo-keto reductase family 1 member B1 |
reviewed |
- |
AKR1B1 |
IC50 |
'=' |
754 |
nM |
| P06746 |
BXGT006496 |
DNA polymerase beta |
reviewed |
Enzyme |
POLB |
Potency |
'=' |
7079 |
nM |
| Q965D6 |
BXGT019601 |
3-oxoacyl-acyl-carrier protein reductase |
unreviewed |
- |
fabG |
Ki |
'=' |
1800 |
nM |
| P06280 |
BXGT006433 |
Alpha-galactosidase A |
reviewed |
- |
GLA |
Potency |
'=' |
50118 |
nM |
| P22303 |
BXGT008751 |
Acetylcholinesterase |
reviewed |
Enzyme |
ACHE |
Ki |
'=' |
65800 |
nM |
| Q9NUW8 |
BXGT021219 |
Tyrosyl-DNA phosphodiesterase 1 |
reviewed |
Enzyme |
TDP1 |
Potency |
- |
33498 |
nM |
| O75496 |
BXGT005132 |
Geminin |
reviewed |
- |
GMNN |
Potency |
- |
3662 |
nM |
| Q7ZJM1 |
BXGT017268 |
Integrase |
unreviewed |
- |
pol |
IC50 |
'=' |
25000 |
nM |
| Q9NR56 |
BXGT021179 |
Muscleblind-like protein 1 |
reviewed |
Nucleic acid binding |
MBNL1 |
Potency |
- |
17782 |
nM |
| P0C6X7 |
BXGT007440 |
Replicase polyprotein 1ab |
reviewed |
- |
rep |
IC50 |
'=' |
20000 |
nM |
| P25779 |
BXGT009067 |
Cruzipain |
reviewed |
- |
|
IC50 |
'=' |
155700 |
nM |
| P11388 |
BXGT007697 |
DNA topoisomerase 2-alpha |
reviewed |
- |
TOP2A |
Activity |
'=' |
4 |
- |
| Q7ZJM1 |
BXGT017268 |
Integrase |
unreviewed |
- |
pol |
IC50 |
'=' |
450 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Kcat |
'=' |
4 |
/s |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Kcat |
'=' |
12 |
/s |
| O42713 |
BXGT004563 |
Polyphenol oxidase 2 |
reviewed |
- |
PPO2 |
Inhibition |
'=' |
7 |
% |
| Q99714 |
BXGT019954 |
3-hydroxyacyl-CoA dehydrogenase type-2 |
reviewed |
Enzyme |
HSD17B10 |
Potency |
'=' |
10000 |
nM |
| P51450 |
BXGT010847 |
Nuclear receptor ROR-gamma |
reviewed |
- |
Rorc |
Potency |
'=' |
11220 |
nM |
| P03372 |
BXGT006059 |
Estrogen receptor |
reviewed |
Nuclear receptor |
ESR1 |
Activity |
- |
- |
- |
| Q16665 |
BXGT013653 |
Hypoxia-inducible factor 1-alpha |
reviewed |
Transcription factor |
HIF1A |
Potency |
'=' |
398 |
nM |
| O42713 |
BXGT004563 |
Polyphenol oxidase 2 |
reviewed |
- |
PPO2 |
Inhibition |
'=' |
12 |
% |
| Q9UBT6 |
BXGT021664 |
DNA polymerase kappa |
reviewed |
- |
POLK |
Potency |
- |
14125 |
nM |
| P05067 |
BXGT006306 |
Amyloid-beta precursor protein |
reviewed |
Enzyme modulator |
APP |
Activity |
'=' |
14 |
% |
| P04798 |
BXGT006265 |
Cytochrome P450 1A1 |
reviewed |
Enzyme |
CYP1A1 |
IC50 |
'=' |
1249 |
nM |
| P04637 |
BXGT006243 |
Cellular tumor antigen p53 |
reviewed |
Transcription factor |
TP53 |
Potency |
'=' |
12589 |
nM |
| Q9NUW8 |
BXGT021219 |
Tyrosyl-DNA phosphodiesterase 1 |
reviewed |
Enzyme |
TDP1 |
Potency |
'=' |
19952 |
nM |
| Q9NPH5 |
BXGT021159 |
NADPH oxidase 4 |
reviewed |
Enzyme |
NOX4 |
IC50 |
'=' |
850 |
nM |
| P04062 |
BXGT006140 |
Lysosomal acid glucosylceramidase |
reviewed |
- |
GBA |
Potency |
- |
12589 |
nM |
| P11387 |
BXGT007696 |
DNA topoisomerase 1 |
reviewed |
Enzyme |
TOP1 |
Activity |
'=' |
0 |
- |
| P27338 |
BXGT009172 |
Amine oxidase [flavin-containing] B |
reviewed |
- |
MAOB |
IC50 |
'=' |
59700 |
nM |
| Q03114 |
BXGT012692 |
Cyclin-dependent-like kinase 5 |
reviewed |
- |
Cdk5 |
EC50 |
'>' |
250000 |
nM |
| P81908 |
BXGT011959 |
Cholinesterase |
reviewed |
- |
BCHE |
Inhibition |
'=' |
0 |
% |
| Q96QE3 |
BXGT019718 |
ATPase family AAA domain-containing protein 5 |
reviewed |
- |
ATAD5 |
Potency |
- |
7304 |
nM |
| P10636 |
BXGT007596 |
Microtubule-associated protein tau |
reviewed |
- |
MAPT |
Potency |
'=' |
17782 |
nM |
| P43405 |
BXGT010386 |
Tyrosine-protein kinase SYK |
reviewed |
Kinase |
SYK |
IC50 |
'=' |
12000 |
nM |
| O42713 |
BXGT004563 |
Polyphenol oxidase 2 |
reviewed |
- |
PPO2 |
Inhibition |
'=' |
33 |
% |
| P42582 |
BXGT010319 |
Homeobox protein Nkx-2.5 |
reviewed |
- |
Nkx2-5 |
IC50 |
'=' |
22000 |
nM |
| Q08369 |
BXGT012983 |
Transcription factor GATA-4 |
reviewed |
- |
Gata4 |
IC50 |
'=' |
22000 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Kcat/Km |
'=' |
0 |
/uM/s |
| Q7ZJM1 |
BXGT017268 |
Integrase |
unreviewed |
- |
pol |
IC50 |
'=' |
32900 |
nM |
| P08170 |
BXGT006736 |
Seed linoleate 13S-lipoxygenase-1 |
reviewed |
- |
LOX1.1 |
Ki |
'=' |
121700 |
nM |
| P08170 |
BXGT006736 |
Seed linoleate 13S-lipoxygenase-1 |
reviewed |
- |
LOX1.1 |
Vmax |
'=' |
0 |
uM |
| P39748 |
BXGT010134 |
Flap endonuclease 1 |
reviewed |
Enzyme |
FEN1 |
Potency |
- |
11220 |
nM |
| P11712 |
BXGT007738 |
Cytochrome P450 2C9 |
reviewed |
- |
CYP2C9 |
Potency |
'=' |
31622 |
nM |
| P14679 |
BXGT008039 |
Tyrosinase |
reviewed |
Enzyme |
TYR |
ID50 |
- |
- |
- |
| P14916 |
BXGT008066 |
Urease subunit alpha |
reviewed |
- |
ureA |
IC50 |
'=' |
35400 |
nM |
| P69996 |
BXGT011656 |
Urease subunit beta |
reviewed |
- |
ureB |
IC50 |
'=' |
35400 |
nM |
| P06239 |
BXGT006427 |
Tyrosine-protein kinase Lck |
reviewed |
Kinase |
LCK |
IC50 |
'=' |
10 |
ug.mL-1 |
| P23977 |
BXGT008936 |
Sodium-dependent dopamine transporter |
reviewed |
- |
Slc6a3 |
EC50 |
'=' |
1450 |
nM |
| P46063 |
BXGT010501 |
ATP-dependent DNA helicase Q1 |
reviewed |
Enzyme |
RECQL |
Potency |
'=' |
8912 |
nM |
| Q03164 |
BXGT012700 |
Histone-lysine N-methyltransferase 2A |
reviewed |
- |
KMT2A |
Potency |
'=' |
35481 |
nM |
| O00255 |
BXGT003903 |
Menin |
reviewed |
- |
MEN1 |
Potency |
'=' |
35481 |
nM |
| Q9UNA4 |
BXGT021785 |
DNA polymerase iota |
reviewed |
- |
POLI |
Potency |
- |
56234 |
nM |
| O95149 |
BXGT005350 |
Snurportin-1 |
reviewed |
- |
SNUPN |
Potency |
- |
50118 |
nM |
| Q14974 |
BXGT013516 |
Importin subunit beta-1 |
reviewed |
Transporter |
KPNB1 |
Potency |
- |
50118 |
nM |
| P62826 |
BXGT011463 |
GTP-binding nuclear protein Ran |
reviewed |
Enzyme modulator |
RAN |
Potency |
- |
50118 |
nM |
| P46063 |
BXGT010501 |
ATP-dependent DNA helicase Q1 |
reviewed |
Enzyme |
RECQL |
Potency |
- |
3162 |
nM |
| P51450 |
BXGT010847 |
Nuclear receptor ROR-gamma |
reviewed |
- |
Rorc |
Potency |
'=' |
17782 |
nM |
| Q965D5 |
BXGT019600 |
Enoyl-acyl-carrier protein reductase |
unreviewed |
- |
fabI |
IC50 |
'=' |
2000 |
nM |
| O75874 |
BXGT005157 |
Isocitrate dehydrogenase [NADP] cytoplasmic |
reviewed |
- |
IDH1 |
Activity |
'=' |
96 |
% |
| P27695 |
BXGT009199 |
DNA-(apurinic or apyrimidinic site) lyase |
reviewed |
- |
APEX1 |
Potency |
- |
4466 |
nM |
| Q9HC97 |
BXGT020527 |
G-protein coupled receptor 35 |
reviewed |
G-protein coupled receptor |
GPR35 |
Activity |
- |
- |
- |
| O15648 |
BXGT004185 |
ATP-dependent 6-phosphofructokinase |
reviewed |
- |
pfk |
Potency |
- |
13459 |
nM |
| P18054 |
BXGT008370 |
Arachidonate 12-lipoxygenase, 12S-type |
reviewed |
Enzyme |
ALOX12 |
Potency |
'=' |
2511 |
nM |
| Q99700 |
BXGT019949 |
Ataxin-2 |
reviewed |
Nucleic acid binding |
ATXN2 |
Potency |
- |
11220 |
nM |
| P0AEK4 |
BXGT007320 |
Enoyl-[acyl-carrier-protein] reductase [NADH] FabI |
reviewed |
- |
fabI |
Activity |
- |
- |
- |
| P11387 |
BXGT007696 |
DNA topoisomerase 1 |
reviewed |
Enzyme |
TOP1 |
Fluorescence intensity |
'=' |
52 |
% |
| P00591 |
BXGT005592 |
Pancreatic triacylglycerol lipase |
reviewed |
- |
PNLIP |
Inhibition |
- |
- |
- |
| Q9UIF8 |
BXGT021718 |
Bromodomain adjacent to zinc finger domain protein 2B |
reviewed |
- |
BAZ2B |
Potency |
- |
79432 |
nM |
| P08254 |
BXGT006758 |
Stromelysin-1 |
reviewed |
Enzyme |
MMP3 |
IC50 |
'=' |
16200 |
nM |
| P03070 |
BXGT006016 |
Large T antigen |
reviewed |
- |
|
EC50 |
- |
47930 |
nM |
| B4URF0 |
BXGT002146 |
Neuraminidase |
unreviewed |
- |
NA |
IC50 |
'=' |
11000 |
nM |
| Q13526 |
BXGT013369 |
Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 |
reviewed |
- |
PIN1 |
Potency |
- |
63095 |
nM |
| O42713 |
BXGT004563 |
Polyphenol oxidase 2 |
reviewed |
- |
PPO2 |
Inhibition |
- |
- |
- |
| P23219 |
BXGT008837 |
Prostaglandin G/H synthase 1 |
reviewed |
Enzyme |
PTGS1 |
Inhibition |
'=' |
75 |
% |
| Q16637 |
BXGT013646 |
Survival motor neuron protein |
reviewed |
Epigenetic regulator |
SMN1 |
Potency |
'=' |
8912 |
nM |
| O42713 |
BXGT004563 |
Polyphenol oxidase 2 |
reviewed |
- |
PPO2 |
Inhibition |
'=' |
6 |
% |
| P35354 |
BXGT009816 |
Prostaglandin G/H synthase 2 |
reviewed |
Enzyme |
PTGS2 |
Inhibition |
'<' |
30 |
% |
| P51450 |
BXGT010847 |
Nuclear receptor ROR-gamma |
reviewed |
- |
Rorc |
Potency |
'=' |
35481 |
nM |
| P49841 |
BXGT010750 |
Glycogen synthase kinase-3 beta |
reviewed |
Kinase |
GSK3B |
IC50 |
'=' |
800 |
nM |
| P49840 |
BXGT010749 |
Glycogen synthase kinase-3 alpha |
reviewed |
Kinase |
GSK3A |
IC50 |
'=' |
800 |
nM |
| P47989 |
BXGT010602 |
Xanthine dehydrogenase/oxidase |
reviewed |
Enzyme |
XDH |
IC50 |
'=' |
550 |
nM |
| Q9WUL0 |
BXGT021941 |
DNA topoisomerase 1 |
reviewed |
- |
Top1 |
IC50 |
'=' |
660 |
nM |
| P38398 |
BXGT010037 |
Breast cancer type 1 susceptibility protein |
reviewed |
Enzyme |
BRCA1 |
Potency |
- |
35481 |
nM |
| P00760 |
BXGT005644 |
Cationic trypsin |
reviewed |
- |
|
IC50 |
'=' |
35300 |
nM |
| Q00534 |
BXGT012533 |
Cyclin-dependent kinase 6 |
reviewed |
Kinase |
CDK6 |
IC50 |
'>' |
300000 |
nM |
| P27695 |
BXGT009199 |
DNA-(apurinic or apyrimidinic site) lyase |
reviewed |
- |
APEX1 |
Potency |
- |
3981 |
nM |
| P10253 |
BXGT007550 |
Lysosomal alpha-glucosidase |
reviewed |
Enzyme |
GAA |
Inhibition |
'=' |
99 |
% |
| Q96QE3 |
BXGT019718 |
ATPase family AAA domain-containing protein 5 |
reviewed |
- |
ATAD5 |
Potency |
- |
9196 |
nM |
| Q04206 |
BXGT012754 |
Transcription factor p65 |
reviewed |
Transcription factor |
RELA |
Inhibition |
- |
- |
- |
| P19838 |
BXGT008526 |
Nuclear factor NF-kappa-B p105 subunit |
reviewed |
Transcription factor |
NFKB1 |
Inhibition |
- |
- |
- |
| Q00653 |
BXGT012541 |
Nuclear factor NF-kappa-B p100 subunit |
reviewed |
Transcription factor |
NFKB2 |
Inhibition |
- |
- |
- |
| O60218 |
BXGT004858 |
Aldo-keto reductase family 1 member B10 |
reviewed |
Enzyme |
AKR1B10 |
Ratio IC50 |
'=' |
10 |
- |
| Q13951 |
BXGT013407 |
Core-binding factor subunit beta |
reviewed |
- |
CBFB |
Potency |
- |
11220 |
nM |
| Q01196 |
BXGT012575 |
Runt-related transcription factor 1 |
reviewed |
Transcription factor |
RUNX1 |
Potency |
- |
11220 |
nM |
| P05067 |
BXGT006306 |
Amyloid-beta precursor protein |
reviewed |
Enzyme modulator |
APP |
Inhibition |
- |
- |
- |
| Q03164 |
BXGT012700 |
Histone-lysine N-methyltransferase 2A |
reviewed |
- |
KMT2A |
Potency |
'=' |
19952 |
nM |
| O00255 |
BXGT003903 |
Menin |
reviewed |
- |
MEN1 |
Potency |
'=' |
19952 |
nM |
| P54132 |
BXGT011032 |
Bloom syndrome protein |
reviewed |
- |
BLM |
Potency |
- |
3827 |
nM |
| P43166 |
BXGT010364 |
Carbonic anhydrase 7 |
reviewed |
- |
CA7 |
Ki |
'=' |
4 |
nM |
| P11387 |
BXGT007696 |
DNA topoisomerase 1 |
reviewed |
Enzyme |
TOP1 |
Fluorescence intensity |
'=' |
0 |
- |
| P19784 |
BXGT008513 |
Casein kinase II subunit alpha' |
reviewed |
Kinase |
CSNK2A2 |
IC50 |
'=' |
1000 |
nM |
| P67870 |
BXGT011566 |
Casein kinase II subunit beta |
reviewed |
Kinase |
CSNK2B |
IC50 |
'=' |
1000 |
nM |
| P09874 |
BXGT006934 |
Poly [ADP-ribose] polymerase 1 |
reviewed |
- |
PARP1 |
IC50 |
'=' |
4265 |
nM |
| Q9H2K2 |
BXGT020454 |
Poly [ADP-ribose] polymerase tankyrase-2 |
reviewed |
- |
TNKS2 |
IC50 |
'=' |
1071 |
nM |
| Q9NUW8 |
BXGT021219 |
Tyrosyl-DNA phosphodiesterase 1 |
reviewed |
Enzyme |
TDP1 |
Potency |
'=' |
39810 |
nM |
| P10253 |
BXGT007550 |
Lysosomal alpha-glucosidase |
reviewed |
Enzyme |
GAA |
Potency |
'=' |
25118 |
nM |
| G5EF15 |
BXGT003524 |
Cytoplasmic zinc-finger protein |
unreviewed |
- |
pos-1 |
EC50 |
'=' |
60374 |
nM |
| P04150 |
BXGT006154 |
Glucocorticoid receptor |
reviewed |
Nuclear receptor |
NR3C1 |
EC150 |
'=' |
18 |
uM |
| P10275 |
BXGT007552 |
Androgen receptor |
reviewed |
Nuclear receptor |
AR |
EC150 |
'=' |
18 |
uM |
| P04745 |
BXGT006255 |
Alpha-amylase 1 |
reviewed |
- |
AMY1A |
Inhibition |
'=' |
88 |
% |
| Q9F4F7 |
BXGT020344 |
4'-phosphopantetheinyl transferase ffp |
reviewed |
- |
ffp |
Potency |
'=' |
63095 |
nM |
| P10635 |
BXGT007595 |
Cytochrome P450 2D6 |
reviewed |
- |
CYP2D6 |
Potency |
'=' |
31622 |
nM |
| P35354 |
BXGT009816 |
Prostaglandin G/H synthase 2 |
reviewed |
Enzyme |
PTGS2 |
IC50 |
'<' |
100 |
ug.mL-1 |
| P35354 |
BXGT009816 |
Prostaglandin G/H synthase 2 |
reviewed |
Enzyme |
PTGS2 |
IC50 |
'=' |
2 |
ug.mL-1 |
| P11388 |
BXGT007697 |
DNA topoisomerase 2-alpha |
reviewed |
- |
TOP2A |
Fluorescence intensity |
'=' |
3 |
- |
| P11387 |
BXGT007696 |
DNA topoisomerase 1 |
reviewed |
Enzyme |
TOP1 |
Fluorescence intensity |
'=' |
99 |
% |
| P07824 |
BXGT006682 |
Arginase-1 |
reviewed |
- |
Arg1 |
IC50 |
'=' |
9000 |
nM |
| Q03164 |
BXGT012700 |
Histone-lysine N-methyltransferase 2A |
reviewed |
- |
KMT2A |
Potency |
'=' |
12589 |
nM |
| O00255 |
BXGT003903 |
Menin |
reviewed |
- |
MEN1 |
Potency |
'=' |
12589 |
nM |
| P04637 |
BXGT006243 |
Cellular tumor antigen p53 |
reviewed |
Transcription factor |
TP53 |
Potency |
'=' |
3162 |
nM |
| P10635 |
BXGT007595 |
Cytochrome P450 2D6 |
reviewed |
- |
CYP2D6 |
AC50 |
- |
- |
- |
| P05177 |
BXGT006336 |
Cytochrome P450 1A2 |
reviewed |
Enzyme |
CYP1A2 |
AC50 |
'=' |
251 |
nM |
| Q9UNA4 |
BXGT021785 |
DNA polymerase iota |
reviewed |
- |
POLI |
Potency |
- |
3981 |
nM |
| Q16236 |
BXGT013624 |
Nuclear factor erythroid 2-related factor 2 |
reviewed |
Enzyme |
NFE2L2 |
Potency |
- |
20596 |
nM |
| O15296 |
BXGT004148 |
Arachidonate 15-lipoxygenase B |
reviewed |
Enzyme |
ALOX15B |
Potency |
- |
10000 |
nM |
| P27487 |
BXGT009187 |
Dipeptidyl peptidase 4 |
reviewed |
Enzyme |
DPP4 |
IC50 |
'=' |
120 |
nM |
| Q15078 |
BXGT013534 |
Cyclin-dependent kinase 5 activator 1 |
reviewed |
Enzyme modulator |
CDK5R1 |
IC50 |
'=' |
3800 |
nM |
| Q00535 |
BXGT012534 |
Cyclin-dependent-like kinase 5 |
reviewed |
Kinase |
CDK5 |
IC50 |
'=' |
3800 |
nM |
| P11388 |
BXGT007697 |
DNA topoisomerase 2-alpha |
reviewed |
- |
TOP2A |
Fluorescence intensity |
'=' |
85 |
% |
| P11388 |
BXGT007697 |
DNA topoisomerase 2-alpha |
reviewed |
- |
TOP2A |
Fluorescence intensity |
'=' |
5 |
- |
| P19532 |
BXGT026110 |
Transcription factor E3 |
reviewed |
- |
TFE3 |
Inhibition |
- |
- |
- |
| B5U2Z5 |
BXGT002179 |
Phenol oxidase |
unreviewed |
- |
|
Activity |
'=' |
45 |
% |
| P39748 |
BXGT010134 |
Flap endonuclease 1 |
reviewed |
Enzyme |
FEN1 |
Potency |
- |
15848 |
nM |
| P00352 |
BXGT005510 |
Retinal dehydrogenase 1 |
reviewed |
Enzyme |
ALDH1A1 |
Potency |
'=' |
8912 |
nM |
| P03468 |
BXGT006079 |
Neuraminidase |
reviewed |
- |
NA |
logIC50 |
'=' |
0 |
- |
| P33261 |
BXGT009707 |
Cytochrome P450 2C19 |
reviewed |
- |
CYP2C19 |
Potency |
'=' |
15848 |
nM |
| P45452 |
BXGT010462 |
Collagenase 3 |
reviewed |
Enzyme |
MMP13 |
Inhibition |
'>' |
30 |
% |
| P37238 |
BXGT009974 |
Peroxisome proliferator-activated receptor gamma |
reviewed |
- |
Pparg |
Emax |
'=' |
7 |
- |
| P37238 |
BXGT009974 |
Peroxisome proliferator-activated receptor gamma |
reviewed |
- |
Pparg |
EC50 |
'=' |
2300 |
nM |
| P03956 |
BXGT006120 |
Interstitial collagenase |
reviewed |
Enzyme |
MMP1 |
IC50 |
- |
- |
- |
| P08170 |
BXGT006736 |
Seed linoleate 13S-lipoxygenase-1 |
reviewed |
- |
LOX1.1 |
Km |
'=' |
148500 |
nM |
| P27169 |
BXGT009161 |
Serum paraoxonase/arylesterase 1 |
reviewed |
- |
PON1 |
KSV |
'=' |
1 |
10'5/M |
| O94782 |
BXGT005332 |
Ubiquitin carboxyl-terminal hydrolase 1 |
reviewed |
Enzyme |
USP1 |
Potency |
- |
44668 |
nM |
| P27695 |
BXGT009199 |
DNA-(apurinic or apyrimidinic site) lyase |
reviewed |
- |
APEX1 |
Potency |
- |
5623 |
nM |
| P28907 |
BXGT009301 |
ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 |
reviewed |
Enzyme |
CD38 |
Ratio IC50 |
'=' |
1 |
- |
| B5U2Z5 |
BXGT002179 |
Phenol oxidase |
unreviewed |
- |
|
IC50 |
'=' |
470000 |
nM |
| B5U2Z5 |
BXGT002179 |
Phenol oxidase |
unreviewed |
- |
|
Activity |
'=' |
67 |
% |
| B5U2Z5 |
BXGT002179 |
Phenol oxidase |
unreviewed |
- |
|
Ki |
'=' |
13110000 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Activity |
'=' |
82 |
nM/s |
| P53355 |
BXGT010985 |
Death-associated protein kinase 1 |
reviewed |
Kinase |
DAPK1 |
IC50 |
'=' |
21000 |
nM |
| Q16678 |
BXGT013656 |
Cytochrome P450 1B1 |
reviewed |
Enzyme |
CYP1B1 |
Ki |
'=' |
56 |
nM |
| O15296 |
BXGT004148 |
Arachidonate 15-lipoxygenase B |
reviewed |
Enzyme |
ALOX15B |
Potency |
- |
15848 |
nM |
| O75496 |
BXGT005132 |
Geminin |
reviewed |
- |
GMNN |
Potency |
- |
18 |
nM |
| P84022 |
BXGT012027 |
Mothers against decapentaplegic homolog 3 |
reviewed |
Transcription factor |
SMAD3 |
Potency |
- |
11220 |
nM |
| P10415 |
BXGT007567 |
Apoptosis regulator Bcl-2 |
reviewed |
Signaling |
BCL2 |
Inhibition |
'=' |
46 |
% |
| Q9UNQ0 |
BXGT021792 |
ATP-binding cassette sub-family G member 2 |
reviewed |
Transporter |
ABCG2 |
IC50 |
'=' |
8900 |
nM |
| O95271 |
BXGT005369 |
Poly [ADP-ribose] polymerase tankyrase-1 |
reviewed |
- |
TNKS |
IC50 |
'=' |
2398 |
nM |
| Q9HC97 |
BXGT020527 |
G-protein coupled receptor 35 |
reviewed |
G-protein coupled receptor |
GPR35 |
Ratio |
'=' |
1 |
- |
| P04798 |
BXGT006265 |
Cytochrome P450 1A1 |
reviewed |
Enzyme |
CYP1A1 |
Ki |
'=' |
890 |
nM |
| P00918 |
BXGT005690 |
Carbonic anhydrase 2 |
reviewed |
- |
CA2 |
Ki |
'=' |
3691 |
nM |
| Q86U86 |
BXGT017611 |
Protein polybromo-1 |
reviewed |
Epigenetic regulator |
PBRM1 |
Kd |
'=' |
12900 |
nM |
| Q9UBT6 |
BXGT021664 |
DNA polymerase kappa |
reviewed |
- |
POLK |
Potency |
- |
1500 |
nM |
| O95398 |
BXGT005378 |
Rap guanine nucleotide exchange factor 3 |
reviewed |
Enzyme modulator |
RAPGEF3 |
Potency |
- |
44668 |
nM |
| O94782 |
BXGT005332 |
Ubiquitin carboxyl-terminal hydrolase 1 |
reviewed |
Enzyme |
USP1 |
Potency |
- |
35481 |
nM |
| Q9HC97 |
BXGT020527 |
G-protein coupled receptor 35 |
reviewed |
G-protein coupled receptor |
GPR35 |
Efficacy |
'=' |
68 |
picometer |
| O95271 |
BXGT005369 |
Poly [ADP-ribose] polymerase tankyrase-1 |
reviewed |
- |
TNKS |
IC50 |
'=' |
2400 |
nM |
| P11021 |
BXGT007644 |
Endoplasmic reticulum chaperone BiP |
reviewed |
- |
HSPA5 |
Potency |
- |
12589 |
nM |
| Q9UBT6 |
BXGT021664 |
DNA polymerase kappa |
reviewed |
- |
POLK |
Potency |
- |
266 |
nM |
| P11309 |
BXGT007685 |
Serine/threonine-protein kinase pim-1 |
reviewed |
Kinase |
PIM1 |
Activity |
'=' |
14 |
% |
| P39900 |
BXGT010148 |
Macrophage metalloelastase |
reviewed |
Enzyme |
MMP12 |
IC50 |
'=' |
980 |
nM |
| P08253 |
BXGT006757 |
72 kDa type IV collagenase |
reviewed |
Enzyme |
MMP2 |
Inhibition |
'>' |
30 |
% |
| P08254 |
BXGT006758 |
Stromelysin-1 |
reviewed |
Enzyme |
MMP3 |
Inhibition |
'>' |
30 |
% |
| P39900 |
BXGT010148 |
Macrophage metalloelastase |
reviewed |
Enzyme |
MMP12 |
Inhibition |
'>' |
30 |
% |
| O94925 |
BXGT005342 |
Glutaminase kidney isoform, mitochondrial |
reviewed |
Enzyme |
GLS |
Potency |
- |
35481 |
nM |
| P46063 |
BXGT010501 |
ATP-dependent DNA helicase Q1 |
reviewed |
Enzyme |
RECQL |
Potency |
'=' |
7943 |
nM |
| P04150 |
BXGT006154 |
Glucocorticoid receptor |
reviewed |
Nuclear receptor |
NR3C1 |
Activity |
- |
- |
- |
| P45452 |
BXGT010462 |
Collagenase 3 |
reviewed |
Enzyme |
MMP13 |
IC50 |
'=' |
10050 |
nM |
| O14763 |
BXGT004093 |
Tumor necrosis factor receptor superfamily member 10B |
reviewed |
- |
TNFRSF10B |
Activity |
- |
- |
- |
| Q14191 |
BXGT013452 |
Werner syndrome ATP-dependent helicase |
reviewed |
Enzyme |
WRN |
Potency |
- |
10000 |
nM |
| Q9Y3R4 |
BXGT022265 |
Sialidase-2 |
reviewed |
Enzyme |
NEU2 |
IC50 |
'=' |
130000 |
nM |
| P46063 |
BXGT010501 |
ATP-dependent DNA helicase Q1 |
reviewed |
Enzyme |
RECQL |
Potency |
'=' |
6309 |
nM |
| P14780 |
BXGT008053 |
Matrix metalloproteinase-9 |
reviewed |
Enzyme |
MMP9 |
IC50 |
'=' |
5620 |
nM |
| O15648 |
BXGT004185 |
ATP-dependent 6-phosphofructokinase |
reviewed |
- |
pfk |
Potency |
- |
10691 |
nM |
| Q99700 |
BXGT019949 |
Ataxin-2 |
reviewed |
Nucleic acid binding |
ATXN2 |
Potency |
- |
19952 |
nM |
| P09917 |
BXGT006941 |
Arachidonate 5-lipoxygenase |
reviewed |
Enzyme |
ALOX5 |
IC50 |
'=' |
1600 |
nM |
| P04798 |
BXGT006265 |
Cytochrome P450 1A1 |
reviewed |
Enzyme |
CYP1A1 |
Ki |
'=' |
2400 |
nM |
| P49841 |
BXGT010750 |
Glycogen synthase kinase-3 beta |
reviewed |
Kinase |
GSK3B |
IC50 |
'=' |
1500 |
nM |
| P11712 |
BXGT007738 |
Cytochrome P450 2C9 |
reviewed |
- |
CYP2C9 |
AC50 |
- |
- |
- |
| O42275 |
BXGT004559 |
Acetylcholinesterase |
reviewed |
- |
ache |
Inhibition |
'=' |
13 |
% |
| P08246 |
BXGT006756 |
Neutrophil elastase |
reviewed |
Enzyme |
ELANE |
IC50 |
'=' |
12700 |
nM |
| P36888 |
BXGT009938 |
Receptor-type tyrosine-protein kinase FLT3 |
reviewed |
Kinase |
FLT3 |
IC50 |
'=' |
830 |
nM |
| Q9HC97 |
BXGT020527 |
G-protein coupled receptor 35 |
reviewed |
G-protein coupled receptor |
GPR35 |
EC50 |
'=' |
3200 |
nM |
| Q9HC97 |
BXGT020527 |
G-protein coupled receptor 35 |
reviewed |
G-protein coupled receptor |
GPR35 |
EC50 |
'=' |
7240 |
nM |
| P05067 |
BXGT006306 |
Amyloid-beta precursor protein |
reviewed |
Enzyme modulator |
APP |
IC50 |
'=' |
6400 |
nM |
| Q13951 |
BXGT013407 |
Core-binding factor subunit beta |
reviewed |
- |
CBFB |
Potency |
- |
10000 |
nM |
| Q01196 |
BXGT012575 |
Runt-related transcription factor 1 |
reviewed |
Transcription factor |
RUNX1 |
Potency |
- |
10000 |
nM |
| Q7ZJM1 |
BXGT017268 |
Integrase |
unreviewed |
- |
pol |
IC50 |
'>' |
20000 |
nM |
| O15648 |
BXGT004185 |
ATP-dependent 6-phosphofructokinase |
reviewed |
- |
pfk |
Potency |
- |
23934 |
nM |
| Q7ZJM1 |
BXGT017268 |
Integrase |
unreviewed |
- |
pol |
IC50 |
'=' |
8900 |
nM |
| P09874 |
BXGT006934 |
Poly [ADP-ribose] polymerase 1 |
reviewed |
- |
PARP1 |
IC50 |
'=' |
4200 |
nM |
| P51570 |
BXGT010858 |
Galactokinase |
reviewed |
Kinase |
GALK1 |
Potency |
'=' |
47269 |
nM |
| Q99714 |
BXGT019954 |
3-hydroxyacyl-CoA dehydrogenase type-2 |
reviewed |
Enzyme |
HSD17B10 |
Potency |
'=' |
15848 |
nM |
| P33261 |
BXGT009707 |
Cytochrome P450 2C19 |
reviewed |
- |
CYP2C19 |
AC50 |
- |
- |
- |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Kcat/Km |
'=' |
0 |
/uM/s |
| P0AEK4 |
BXGT007320 |
Enoyl-[acyl-carrier-protein] reductase [NADH] FabI |
reviewed |
- |
fabI |
Ki |
'=' |
7050 |
nM |
| Q965D7 |
BXGT019602 |
Beta-hydroxyacyl-ACP dehydratase |
unreviewed |
- |
fabZ |
IC50 |
'=' |
5000 |
nM |
| Q07817 |
BXGT012941 |
Bcl-2-like protein 1 |
reviewed |
Signaling |
BCL2L1 |
Inhibition |
'=' |
49 |
% |
| P22748 |
BXGT008802 |
Carbonic anhydrase 4 |
reviewed |
- |
CA4 |
Ki |
'=' |
293 |
nM |
| P02766 |
BXGT005953 |
Transthyretin |
reviewed |
Transporter |
TTR |
Kd |
'=' |
70 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
Ratio IC50 |
'=' |
25 |
- |
| P47989 |
BXGT010602 |
Xanthine dehydrogenase/oxidase |
reviewed |
Enzyme |
XDH |
IC50 |
'=' |
7800 |
nM |
| Q9H2K2 |
BXGT020454 |
Poly [ADP-ribose] polymerase tankyrase-2 |
reviewed |
- |
TNKS2 |
IC50 |
'=' |
1100 |
nM |
| P07363 |
BXGT026132 |
Chemotaxis protein CheA |
reviewed |
- |
cheA |
IC50 |
'=' |
111000 |
nM |
| P27695 |
BXGT009199 |
DNA-(apurinic or apyrimidinic site) lyase |
reviewed |
- |
APEX1 |
Potency |
- |
5011 |
nM |
| O60218 |
BXGT004858 |
Aldo-keto reductase family 1 member B10 |
reviewed |
Enzyme |
AKR1B10 |
IC50 |
'=' |
107800 |
nM |
| P19784 |
BXGT008513 |
Casein kinase II subunit alpha' |
reviewed |
Kinase |
CSNK2A2 |
IC50 |
'<' |
1000 |
nM |
| P68400 |
BXGT011599 |
Casein kinase II subunit alpha |
reviewed |
Kinase |
CSNK2A1 |
IC50 |
'<' |
1000 |
nM |
| P67870 |
BXGT011566 |
Casein kinase II subunit beta |
reviewed |
Kinase |
CSNK2B |
IC50 |
'<' |
1000 |
nM |
| P15121 |
BXGT008083 |
Aldo-keto reductase family 1 member B1 |
reviewed |
Enzyme |
AKR1B1 |
IC50 |
'=' |
3200 |
nM |
| O60218 |
BXGT004858 |
Aldo-keto reductase family 1 member B10 |
reviewed |
Enzyme |
AKR1B10 |
Inhibition |
- |
- |
- |
| P08253 |
BXGT006757 |
72 kDa type IV collagenase |
reviewed |
Enzyme |
MMP2 |
IC50 |
'=' |
3610 |
nM |
| P43405 |
BXGT010386 |
Tyrosine-protein kinase SYK |
reviewed |
Kinase |
SYK |
EC50 |
'=' |
4500 |
nM |
| P10253 |
BXGT007550 |
Lysosomal alpha-glucosidase |
reviewed |
Enzyme |
GAA |
IC50 |
'=' |
5 |
ug.mL-1 |
| P08246 |
BXGT006756 |
Neutrophil elastase |
reviewed |
Enzyme |
ELANE |
Ki |
'=' |
10100 |
nM |
| P16050 |
BXGT008184 |
Arachidonate 15-lipoxygenase |
reviewed |
Enzyme |
ALOX15 |
IC50 |
'=' |
5000 |
nM |
| P80457 |
BXGT011911 |
Xanthine dehydrogenase/oxidase |
reviewed |
- |
XDH |
IC50 |
'=' |
10200 |
nM |
| O43570 |
BXGT004601 |
Carbonic anhydrase 12 |
reviewed |
- |
CA12 |
Ki |
'=' |
60 |
nM |
| O60218 |
BXGT004858 |
Aldo-keto reductase family 1 member B10 |
reviewed |
Enzyme |
AKR1B10 |
IC50 |
'=' |
9200 |
nM |
| P15121 |
BXGT008083 |
Aldo-keto reductase family 1 member B1 |
reviewed |
Enzyme |
AKR1B1 |
IC50 |
'=' |
600 |
nM |
| Q15118 |
BXGT013540 |
[Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 1, mitochondrial |
reviewed |
Kinase |
PDK1 |
Inhibition |
'=' |
31 |
% |
| P05067 |
BXGT006306 |
Amyloid-beta precursor protein |
reviewed |
Enzyme modulator |
APP |
Inhibition |
'=' |
86 |
% |
| P03070 |
BXGT006016 |
Large T antigen |
reviewed |
- |
|
IC50 |
- |
11900 |
nM |
| P07378 |
BXGT006616 |
Phosphoglycerate kinase, glycosomal |
reviewed |
- |
|
Potency |
- |
28183 |
nM |
| P02766 |
BXGT005953 |
Transthyretin |
reviewed |
Transporter |
TTR |
IC50 |
'=' |
7000 |
nM |
| Q86U86 |
BXGT017611 |
Protein polybromo-1 |
reviewed |
Epigenetic regulator |
PBRM1 |
Activity |
- |
- |
- |
| M5KM83 |
BXGT026161 |
Sensor protein kinase WalK family protein |
unreviewed |
- |
PCS8203_00348 |
IC50 |
'=' |
216000 |
nM |
| P47989 |
BXGT010602 |
Xanthine dehydrogenase/oxidase |
reviewed |
Enzyme |
XDH |
Inhibition |
'>' |
50 |
% |